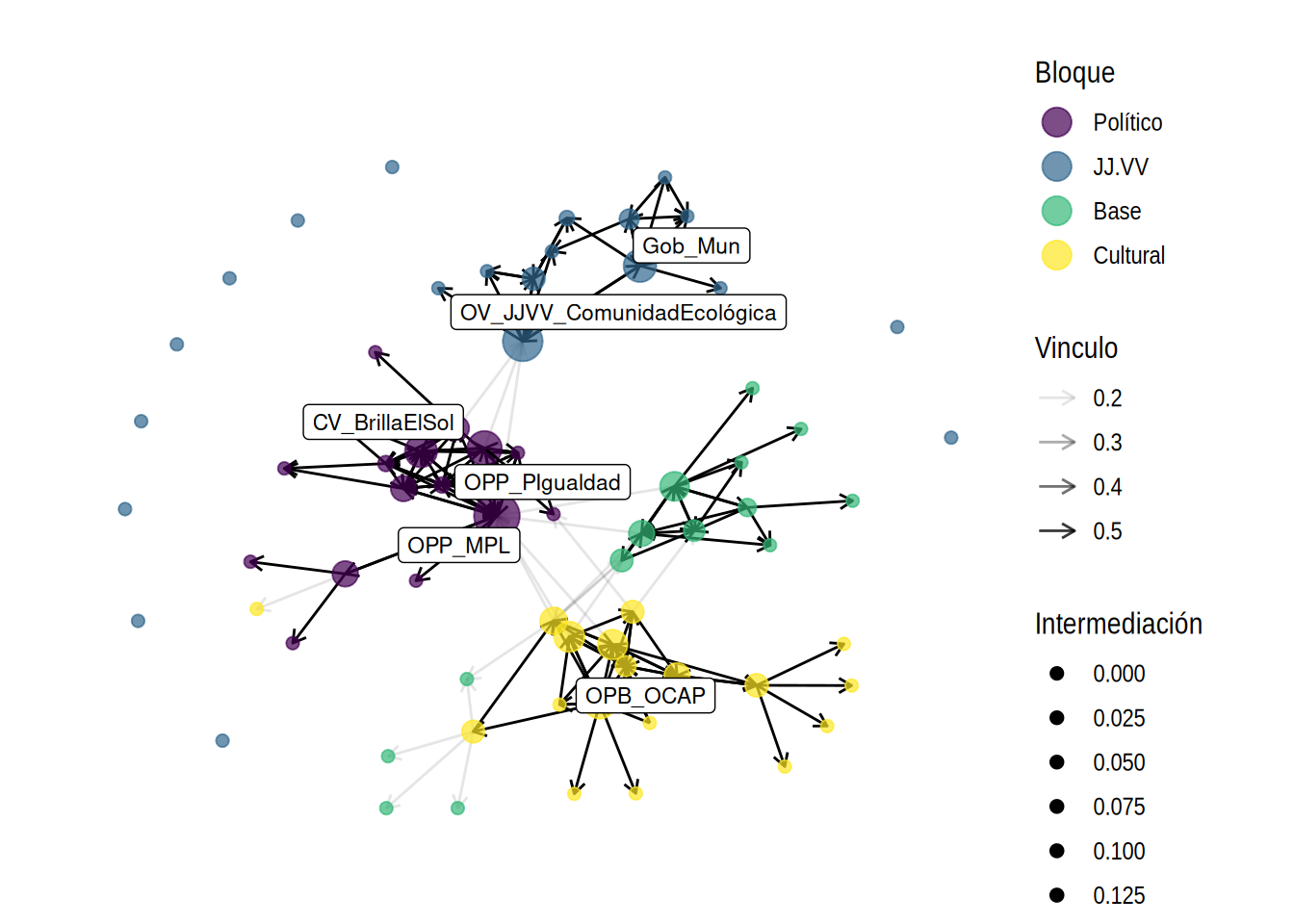
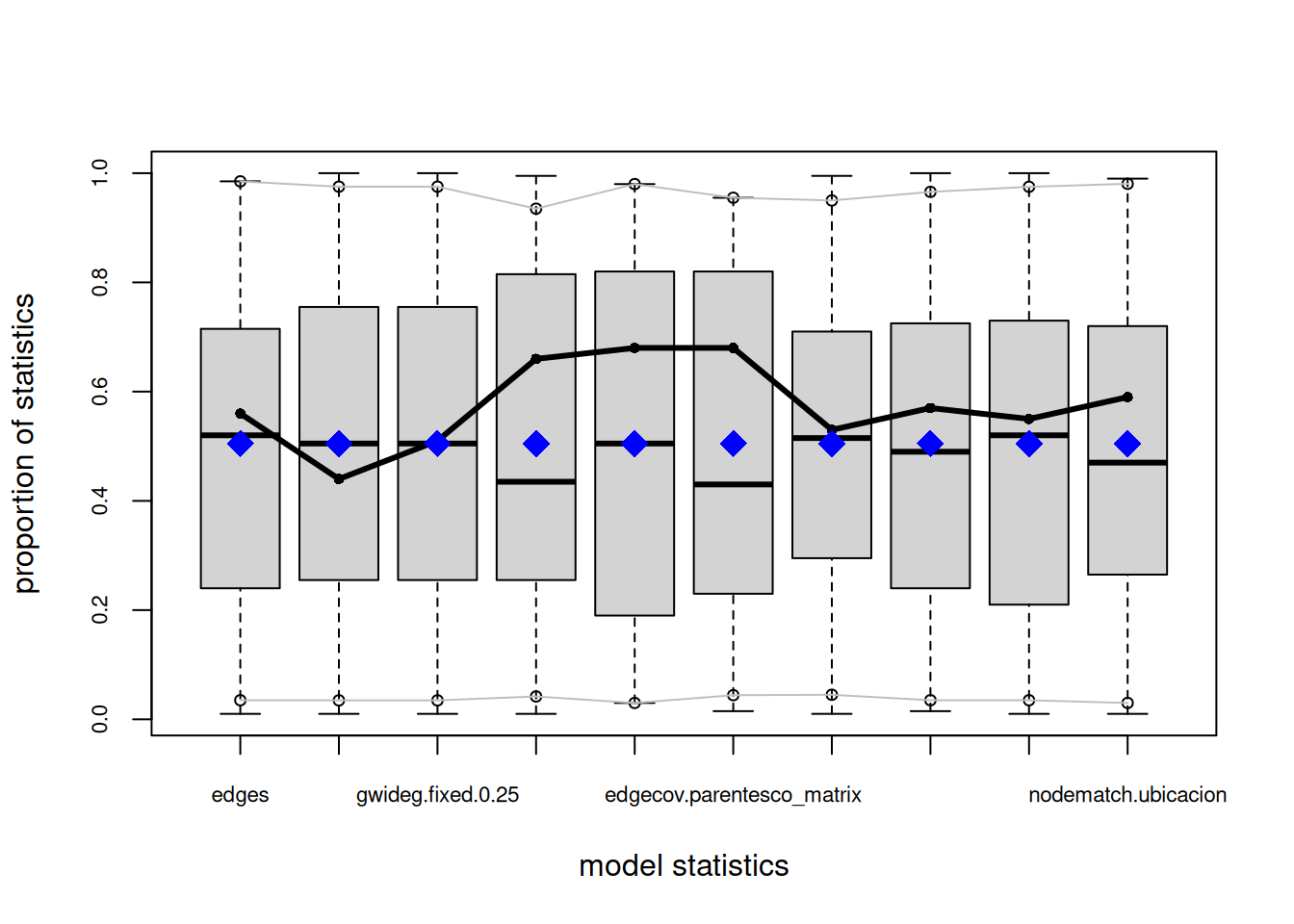
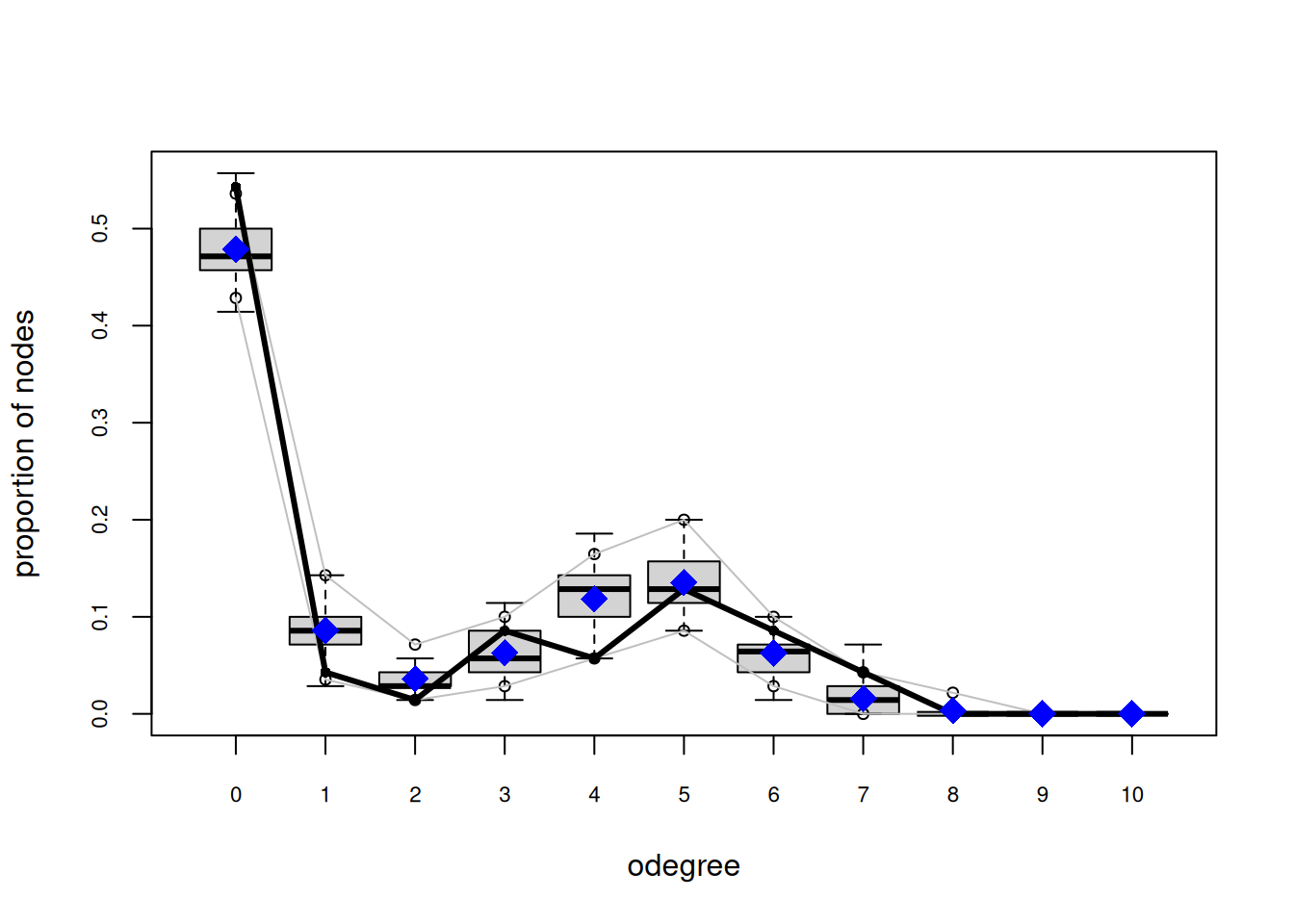
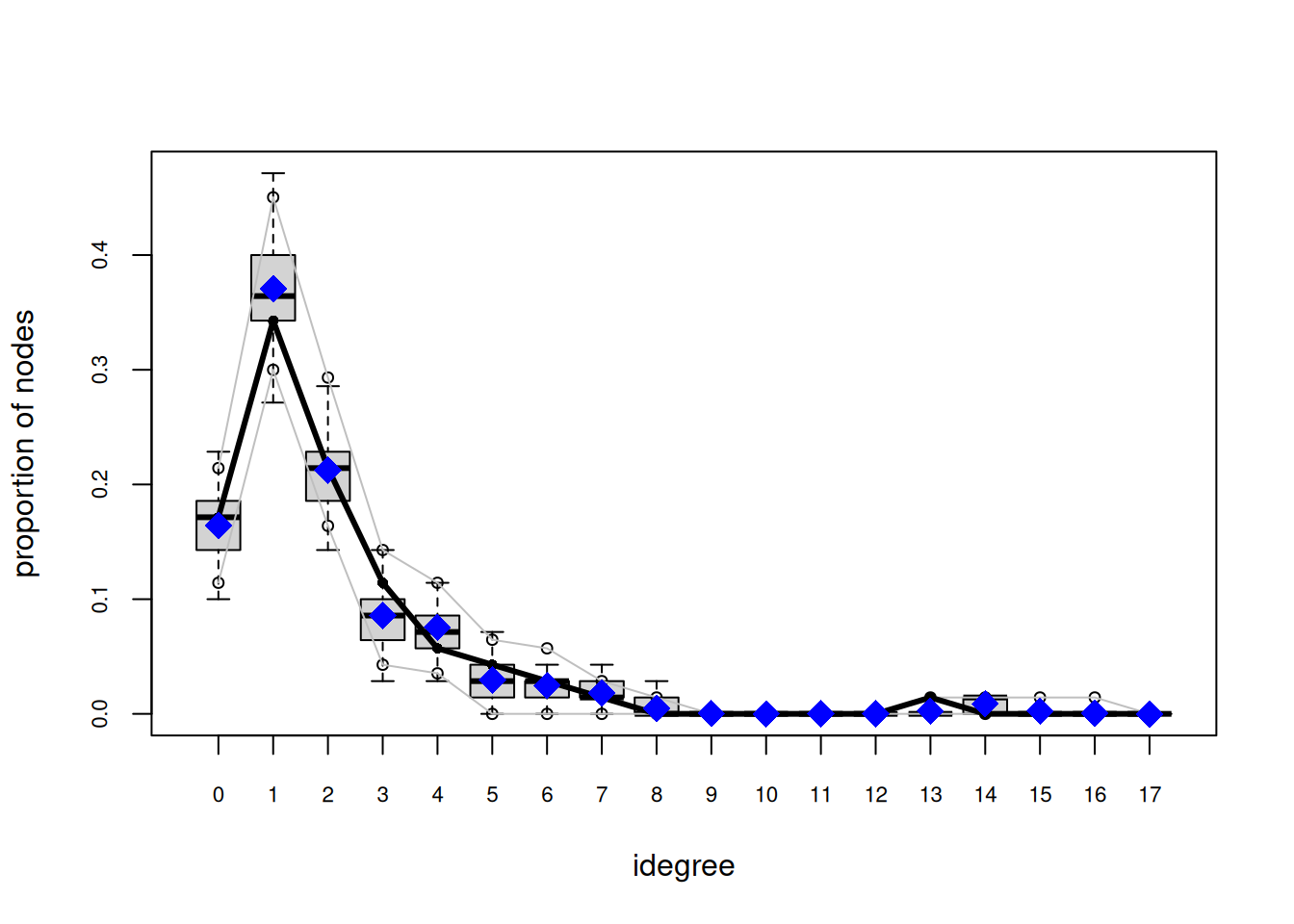
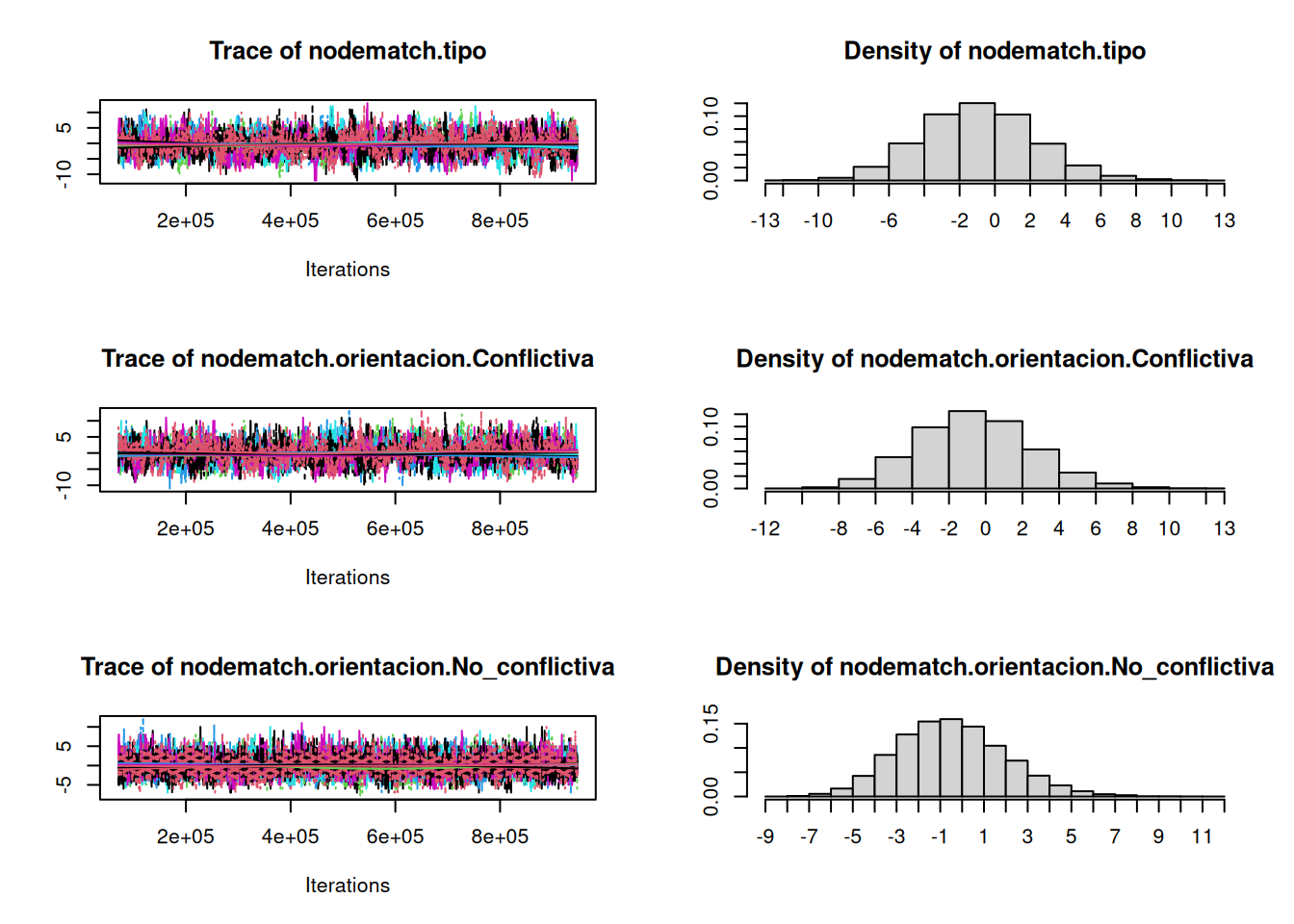
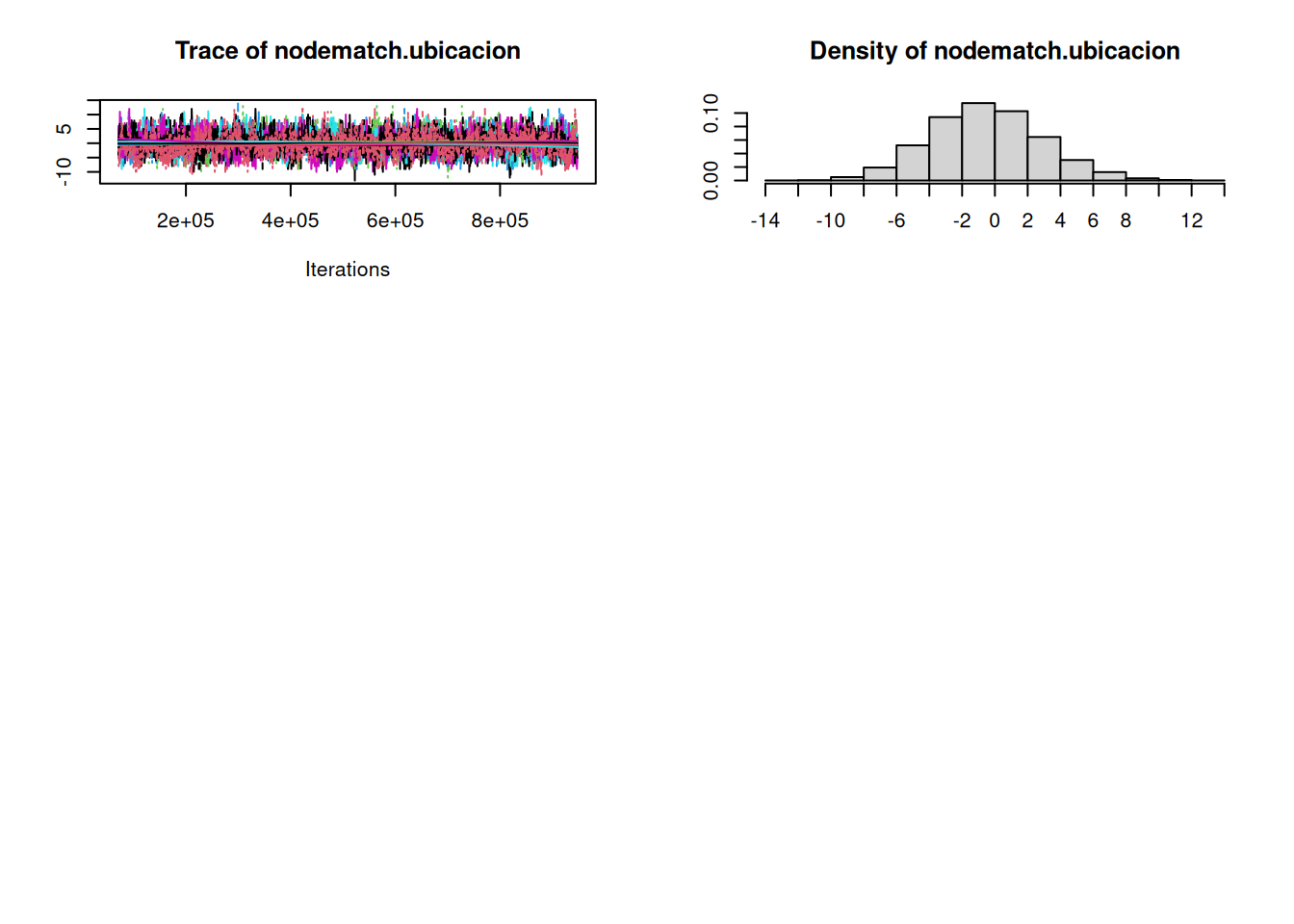
org_attributes$block <- as.factor(org_attributes$`X1PosciónCONCOR`)
improved_diffusion_v9 <- function(networks, attributes, seeds,
max_iterations = 1000,
convergence_threshold = 0.001,
memory_window = 5,
use_heterogeneous_thresholds = TRUE) {
# 1. Umbrales realistas para comportamiento conflictivo
thresholds <- list(
base = 0.05, # Umbral base alto para comportamiento conflictivo
by_type = c(
" Club deportivo " = 0.15,
" Comité de vivienda " = 0.06,
" Organización cultural " = 0.10,
" Organización política de base " = 0.07,
" Organización política de pobladores " = 0.05,
" Organización vecinal " = 0.10,
"Otros" = 0.15
))
# 2. Validación de entrada y preprocesamiento
networks <- lapply(networks, function(x) {
if(!is.matrix(x)) stop("Todas las redes deben ser matrices")
return(as.matrix(x))
})
if(!all(sapply(networks, function(x) all(dim(x) == dim(networks[[1]])))))
stop("Todas las redes deben tener las mismas dimensiones")
# 3. Inicialización
n_nodes <- nrow(networks[[1]])
weights <- c(0.35, 0.35, 0.15, 0.15) # Más peso a confianza y valores
# 4. Crear red compuesta
composite_net <- Reduce("+", Map("*", networks, weights))
# 5. Calcular medidas estructurales
g <- igraph::graph_from_adjacency_matrix(composite_net,
weighted = TRUE,
mode = "directed")
# 6. Medidas de centralidad
centrality_measures <- list(
between = igraph::betweenness(g, normalized = TRUE),
degree = igraph::degree(g, mode = "all", normalized = TRUE),
eigen = igraph::eigen_centrality(g)$vector,
authority = igraph::authority_score(g)$vector
)
# 7. Identificación de roles estructurales
role_scores <- data.frame(
node = 1:n_nodes,
between_score = scale(centrality_measures$between),
degree_score = scale(centrality_measures$degree),
eigen_score = scale(centrality_measures$eigen),
authority_score = scale(centrality_measures$authority)
)
# Calcular scores compuestos
role_scores$broker_score <- role_scores$between_score + role_scores$degree_score
role_scores$core_score <- role_scores$eigen_score + role_scores$authority_score
# Asignar roles con umbrales más estrictos
role_scores$role <- with(role_scores, {
ifelse(broker_score > quantile(broker_score, 0.85), "broker",
ifelse(core_score > quantile(core_score, 0.85), "core",
ifelse(authority_score > quantile(authority_score, 0.85), "authority",
"peripheral")))
})
# 8. Funciones auxiliares mejoradas
calc_local_clustering <- function(node, networks) {
cluster_scores <- sapply(networks, function(x) {
neighbors <- which(x[node,] > 0)
if(length(neighbors) < 2) return(0)
submat <- x[neighbors, neighbors]
sum(submat) / (length(neighbors) * (length(neighbors)-1))
})
weighted.mean(cluster_scores, weights)
}
calc_exposure_time <- function(node, history, window = memory_window) {
# Si no hay historia, retornar 0
if(is.null(history) || is.null(dim(history)) || nrow(history) == 0) return(0)
# Si no hay vecinos, retornar 0
if(length(node_neighbors[[node]]) == 0) return(0)
# Ajustar ventana si es necesario
current_window <- min(window, nrow(history))
# Obtener historia reciente
if(current_window == 1) {
recent_history <- matrix(history, nrow=1)
} else {
recent_history <- tail(history, current_window)
}
# Calcular exposición de vecinos
exposed_neighbors <- rowSums(recent_history[, node_neighbors[[node]], drop=FALSE])
# Calcular pesos
weights <- exp(-(current_window:1)/3)
# Retornar media ponderada
return(weighted.mean(exposed_neighbors, weights))
}
identity_alignment <- function(node, attributes) {
# Mide similitud en atributos organizacionales
same_type <- attributes$tipo == attributes$tipo[node]
same_orientation <- attributes$Orientación == attributes$Orientación[node]
return(mean(same_type & same_orientation))
}
collective_action_cost <- function(node, role_scores) {
# Costo base por visibilidad estructural
base_cost <- 1 + abs(role_scores$degree_score[node])
# Modificador por rol
role_modifier <- switch(role_scores$role[node],
"broker" = 1.3,
"core" = 1.2,
"authority" = 1.1,
1.0)
return(base_cost * role_modifier)
}
# 9. Inicialización de estados y memorias
states <- rep(0, n_nodes)
states[seeds] <- 1
# Crear lista de vecinos
node_neighbors <- lapply(1:n_nodes, function(i) {
unique(unlist(lapply(networks, function(x) which(x[i,] > 0))))
})
# 10. Inicialización de matrices de historia
history <- matrix(0, nrow = max_iterations, ncol = n_nodes)
history[1,] <- states
exposure_memory <- matrix(0, nrow = max_iterations, ncol = n_nodes)
exposure_memory[1,] <- sapply(1:n_nodes, function(i) {
if(length(node_neighbors[[i]]) > 0) {
mean(states[node_neighbors[[i]]])
} else {
0
}
})
clustering_memory <- matrix(0, nrow = max_iterations, ncol = n_nodes)
clustering_memory[1,] <- sapply(1:n_nodes, function(i) calc_local_clustering(i, networks))
# 11. Proceso de difusión mejorado
for(iter in 2:max_iterations) {
old_states <- states
# Procesar nodos inactivos
inactive_nodes <- which(states == 0)
for(i in inactive_nodes) {
# Calcular influencia base
influence <- 0
total_weight <- 0
# Influencia por tipo de red
for(n in seq_along(networks)) {
neighbors <- which(networks[[n]][i,] > 0)
if(length(neighbors) > 0) {
# Influencia ponderada por tipo de vínculo y tiempo
net_influence <- sum(states[neighbors]) / length(neighbors)
# Factor temporal de exposición
exposure_time <- calc_exposure_time(i, history[1:(iter-1), , drop=FALSE])
temporal_factor <- 1 - exp(-0.05 * exposure_time)
# Efecto de clustering local
cluster_effect <- calc_local_clustering(i, networks)
cluster_multiplier <- 1 + (cluster_effect * 2)
# Efecto de identidad
identity_effect <- identity_alignment(i, attributes)
# Costo de acción colectiva
action_cost <- collective_action_cost(i, role_scores)
# Combinar efectos
combined_influence <- net_influence * weights[n] *
temporal_factor * cluster_multiplier *
(1 + identity_effect) / action_cost
influence <- influence + combined_influence
total_weight <- total_weight + weights[n]
}
}
if(total_weight > 0) {
# Normalizar influencia
influence <- influence / total_weight
if(use_heterogeneous_thresholds) {
# Threshold dinámico con heterogeneidad
base_threshold <- thresholds$base
type_threshold <- thresholds$by_type[attributes$tipo[i]]
} else {
# Threshold dinámico sin heterogeneidad
base_threshold <- thresholds$base
type_threshold <- 1
}
# Ajustar threshold por exposición temporal y clustering
exposure_effect <- mean(exposure_memory[1:max(1,iter-1), i], na.rm=TRUE)
cluster_effect <- mean(clustering_memory[1:max(1,iter-1), i], na.rm=TRUE)
final_threshold <- (base_threshold * type_threshold) *
exp(-0.05 * exposure_effect) *
(1 - 0.2 * cluster_effect)
# Probabilidad de adopción
if(influence >= final_threshold) {
# Fricción en adopción
adoption_prob <- 0.3 * (1 + exposure_effect)
states[i] <- rbinom(1, 1, min(1, adoption_prob))
}
}
# Actualizar memorias
exposure_memory[iter, i] <- ifelse(length(node_neighbors[[i]]) > 0,
sum(states[node_neighbors[[i]]]) / length(node_neighbors[[i]]),
0)
clustering_memory[iter, i] <- calc_local_clustering(i, networks)
}
# Guardar historia
history[iter,] <- states
# Verificar convergencia
if(iter > 20) {
recent_window <- (iter-19):iter
change_rate <- mean(diff(colMeans(history[recent_window,])))
if(abs(change_rate) < convergence_threshold) break
}
if(all(old_states == states) && iter > 10) break
}
# 12. Calcular métricas finales
final_metrics <- list(
adoption_rate = mean(states),
clustering_effect = sapply(1:n_nodes, function(i) calc_local_clustering(i, networks)),
temporal_exposure = colMeans(exposure_memory[1:iter,], na.rm=TRUE),
convergence_iteration = iter,
change_trajectory = rowMeans(history[1:iter,]),
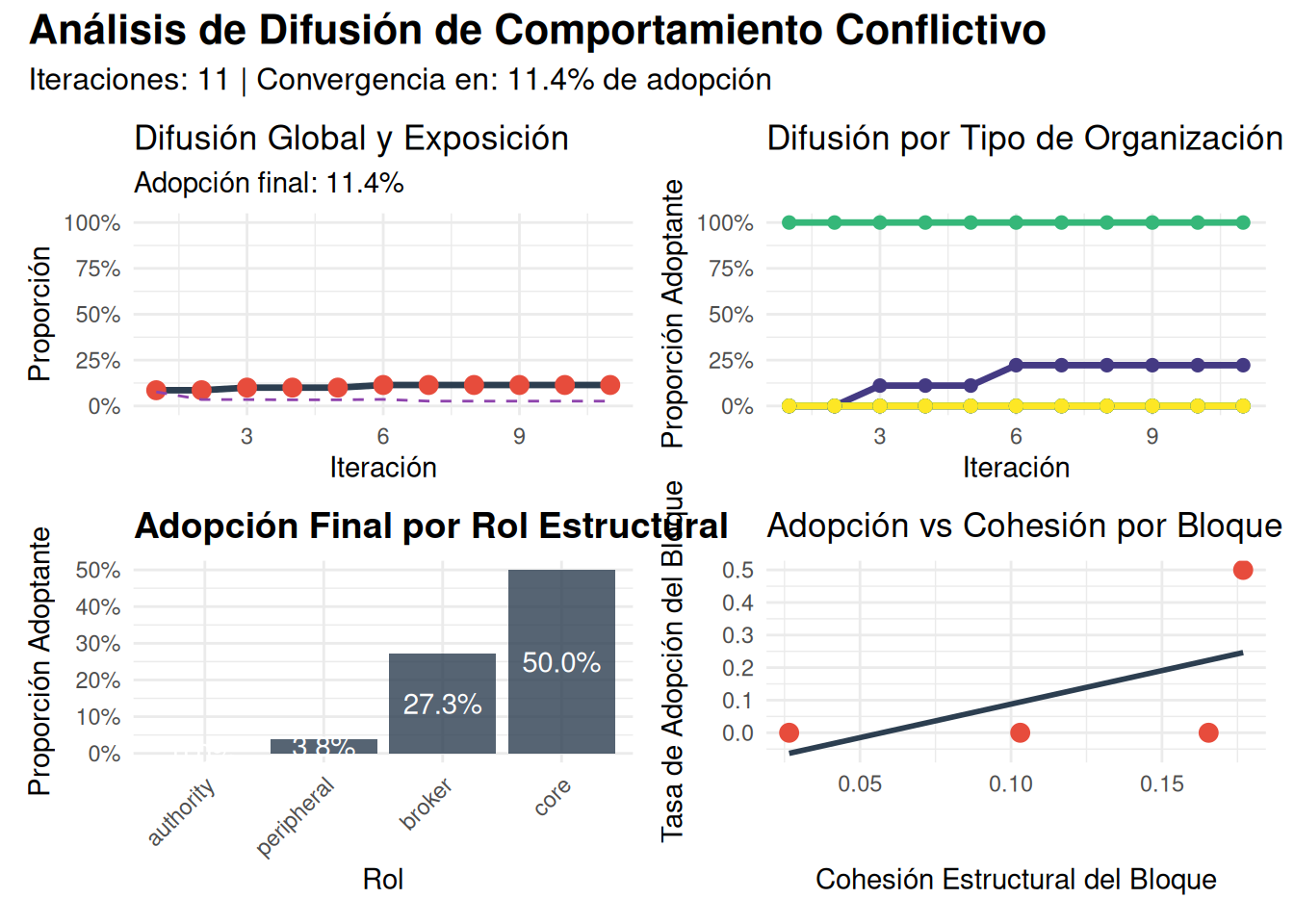
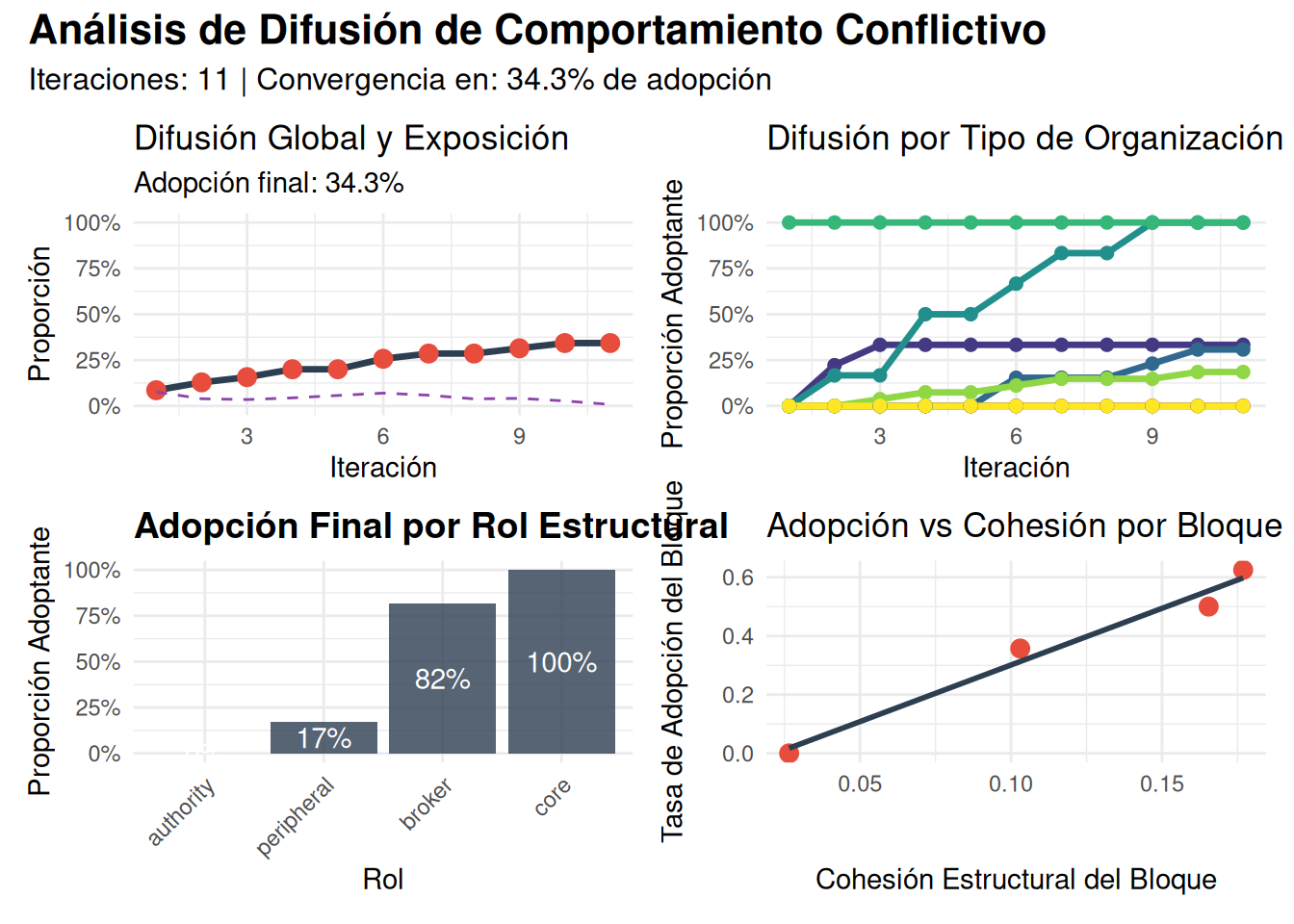
adoption_by_type = tapply(states, attributes$tipo, mean),
adoption_by_role = tapply(states, role_scores$role, mean),
final_thresholds = sapply(1:n_nodes, function(i) {
base_threshold <- if(use_heterogeneous_thresholds) {
thresholds$by_type[attributes$tipo[i]]
} else {
thresholds$base
}
exposure_effect <- mean(exposure_memory[1:iter, i], na.rm=TRUE)
cluster_effect <- mean(clustering_memory[1:iter, i], na.rm=TRUE)
base_threshold * exp(-0.05 * exposure_effect) *
(1 - 0.2 * cluster_effect)
}),
network_effects = list(
clustering = mean(sapply(1:n_nodes, function(i) calc_local_clustering(i, networks))),
exposure = mean(colMeans(exposure_memory[1:iter,], na.rm=TRUE)),
identity = mean(sapply(1:n_nodes, function(i) identity_alignment(i, attributes))),
costs = mean(sapply(1:n_nodes, function(i) collective_action_cost(i, role_scores)))
)
)
# 13. Retornar resultados
return(list(
final_states = states,
history = history[1:iter,],
n_iterations = iter,
role_scores = role_scores,
centrality_measures = centrality_measures,
composite_net = composite_net,
exposure_memory = exposure_memory[1:iter,],
clustering_memory = clustering_memory[1:iter,],
final_metrics = final_metrics,
converged = iter < max_iterations,
parameters = list(
thresholds = thresholds,
weights = weights,
memory_window = memory_window,
convergence_threshold = convergence_threshold,
use_heterogeneous_thresholds = use_heterogeneous_thresholds
)
))
}